vivaxGEN: A global collaboration using translational genomics to develop new molecular surveillance tools to support the elimination of P. vivax
Aim:
To develop novel molecular surveillance tools that provide clinically relevant information on emerging parasite adaptations, transmission dynamics, the major routes of parasite spread within and across borders, and the impact of local treatment policies on the dormant liver-stage reservoir.
The problem – limitations of traditional surveillance tools:
In the Asia-Pacific, Horn of Africa and Americas, Plasmodium vivax is now the predominant cause of malaria, affecting the world’s poorest and most vulnerable communities. Efforts to contain P. vivax have been constrained by the parasite’s ability to adapt and form dormant liver stages that can relapse months after inoculation. Traditional malaria surveillance focuses on measuring the prevalence or incidence of infection, essentially counting clinical cases. Whilst this is useful for monitoring transmission reduction, it does not inform on a dynamic parasite population that may be adapting to public health interventions. New surveillance tools have potential to provide far greater insights into the parasite reservoirs, and how they are adapting and spreading within and across national borders.
Our strategy to develop and implement new molecular surveillance tools:
Our research program incorporates molecular biology, population genetics and genomics, and software development to inform on the biology of P. vivax. To assimilate these data, we are developing laboratory and online analytical tools for molecular surveillance. Our strategy employs a 4-step plan:
- Genomic analyses of a growing global collection of P. vivax infections to identify subsets of markers (barcodes) that can inform on local transmission patterns, parasite spread within and across borders, imported infections and their origins, drug resistance and other forms of parasite adaptation.
- Laboratory-based assays to generate these P. vivax barcode markers to support cost-effective high throughput genotyping of representative host and parasite population.
- Bioinformatics tools to support local researchers, control programs and other key stakeholders to effectively analyse and interpret the parasite genotyping data.
- In country capacity building to ensure effective translation of molecular surveillance tools and generate locally informative data in a timely manner. The program currently comprises partners from 16 countries, that have collectively contributed a global collection of >1,000 P. vivax genomes and >6,000 patient samples from cross-sectional surveys as well as unique longitudinal cohorts with up to one year follow up.
In the next five years, specific objectives of the program are to:
- Expand the global repository of P. vivax genomes and generate new knowledge on emerging molecular adaptations, including evolving drug resistance
- Transfer our earlier microsatellite-based tools into higher-throughput SNP methodologies
- Expand on earlier tools for mapping imported infections by developing higher resolution tools to track P. vivax spread within-country and across border regions
- Develop molecular tools to track relapses that can inform on the efficacy and adherence of local treatment policies to minimize the liver-stage reservoir
- Implement the molecular surveillance tools in a range of endemic settings at sites in Bhutan, China, Indonesia, Malaysia and Vietnam.
Implications for policy and practice:
This program will provide researchers and malaria control programs in endemic countries with actionable knowledge, supporting the rapid containment and elimination of P. vivax.
For further information:
Contact Associate Professor Sarah Auburn via email.
The molecular surveillance program began as a collaborative project among researchers within the Asia Pacific Malaria Elimination Network (APMEN) vivax Working Group in 2010. It has since expanded to a global network of partners from the Asia-Pacific, Horn of Africa, Saharan Africa and Americas.
Where we work
.jpg)
Our work is closely aligned with the Malaria Genomic Epidemiology Network (malariaGEN) SpotMalaria program, employing a similar operational framework.
- Identifying use cases: To ensure that our surveillance tools meet the needs of National Malaria Control Programs (NMCPs), we hold workshops for representatives of NMCPs, the WHO, research partners, and key stakeholders involved in malaria control and elimination programs. The objectives of these workshops are to identify the meaningful use cases so that priority is given to molecular surveillance tools that can support the operational activities of NMCPs. Efforts are then focused on defining the challenges in implementing these tools in country and developing strategies to overcome these.
- Previous Molecular Workshops:
- 2-day parasite surveillance workshop co-funded by APMEN and the Australian DFAT, Jakarta, Indonesia, Nov 2019
- P. vivax genotyping workshop, 5th International Conference on Plasmodium vivax research, Bali, Indonesia, June 2015
- Parasite surveillance session, APMEN Vivax Working Group, Manila, Philippines, March 2014
- 1-day data analysis and sharing workshop, APMEN Vivax Working Group, Incheon, Korea, May 2012
- 1-day parasite genotyping workshop, APMEN Vivax Working Group, Sabah, Malaysia, 2011
- Building a collaborative network: vivaxGEN. Over the past 10 years, we have established partnerships with collaborators from 22 institutes in 16 endemic countries across the globe.
- Whole genome sequencing: Samples that have been collected using suitable processing methods (see our SOP below), are subject to whole genome sequencing at the Wellcome Sanger Institute, UK, as part of a collaboration with the malariaGEN program.
- Marker selection: Bioinformatic analysis of the genomic data to identify suitable barcode markers is undertaken as a collaborative effort between research leads at Menzies, the Eijkman Institute for Molecular Biology, Indonesia, and the Big Data Institute, UK.
- Assay development for the selected barcode markers is being undertaken at the Wellcome Sanger Institute. The development of vivax-specific bioinformatic tools to support local research agendas and data interpretation by NMCPs is being undertaken by researchers at Menzies and the Eijkman Institute. These tools will be aligned with the SpotMalaria bioinformatics pipeline to optimize all-species reporting to local NMCPs.
- In-country implementation of the laboratory assays and informatic tools is being led by Menzies and the Wellcome Sanger Institute.

Project team
Principal investigator:
Chief investigators:
- Professor Ric Price
- Olivo Miotto
- Dominic Kwiatkowski
Menzies investigators:
- Dr Jutta Marfurt
- Angela Rumaseb
- Dr Benedikt Ley
- Dr Kamala Ley-Thriemer
- Dr Matthew Grigg
- Associate Professor Bridget Barber
- Professor Nicholas Anstey
Project managers:
- Catherine Martel
- Cindy Fahey
Endemic country collaborators:
- Asia Pacific Malaria Elimination Network (Global)
- Nangarhar University, Afghanistan
- Awab Ghulam Rahim
- International Centre for Diarrheal Diseases Research, Bangladesh
- Shafiul Alam
- Wasif Khan
- Department of Public Health, Bhutan
- Sonam Wangchuk
- Jiangsu Institute of Parasitic Diseases, China
- Yaobao Liu o Gao Qi
- National Institute of Parasitological Diseases, China
- Duoquan Wang
- Centro Internacionale de Entrenamiento e Investigaciones Medicas, Colombia
- Zuleima Pava
- Diego Echeverry
- Universidad de Antioquia, Colombia
- Tatiana Lopera-Mesa
- Armauer Hansen Research Unit, Ethiopia
- Sisay Alemu
- Endalamaw Gadisa
- Abraham Aseffa
- Addis Ababa University, Ethiopia
- Beyene Petros
- Ethiopian Public Health Institute, Ethiopia
- Ashenafi Assefa
- Eijkman Institute of Molecular Biology, Indonesia
- Hidayat Trimarsanto
- Rintis Noviyanti
- Hormozgan University, Iran
- Yaghoob Hamedi
- National Institute of Health, Korea CDC, Republic of Korea
- Jung-Yeon Kim
- Youn-Kyoung Goo
- Centre for Malaria Parasitology and Entomology, Laos
- Mayfong Mayxay
- Clinical Research Centre, Queen Elizabeth Hospital, Malaysia
- Timothy William
- Tribhuvan University, Nepal
- Prakash Ghimire
- Agha Khan University, Pakistan
- Najia Ghanchi
- University of Khartoum, Sudan
- Ishag Adam
- Shoklo Malaria Research Unit, Thailand
- Francois Nosten
- Mahidol-Oxford Tropical Medicine Research Unit, Thailand
- Nicholas White
- Nicholas Day
- National Institute of Malariology, Parasitology and Entomology, Vietnam
- Xa Nguyen Xuan
- Thuanthi Nguyen
- Oxford University Clinical Research Unit, Vietnam
- Nhien Nguyen Thanh Thuy
- Nguyen Hoang Chau
- Tran Tinh Hien
Whole genome sequencing and assay development support:
- Wellcome Sanger Institute, UK
- Sasha Siegel
- Sonia Goncalves
- Chris Jacobs
- Dominic Kwiatkowski
- Big Data Institute, UK
- Richard Pearson
- Roberto Amato
- Olivo Miotto
- Dominic Kwiatkowski
Funders:
- The Wellcome Trust
- Bill and Melinda Gates Foundation
- Australian National Health and Medical Research Council
- Medical Research Council UK
- Australian Department of Foreign Affairs and Trade
- Australian Centre of Research Excellence in Malaria Elimination
Laboratory SOPs:
- SOP for preparing high-quality patient P. vivax samples for whole genome sequencing
- Paper: Effective preparation of Plasmodium vivax field isolates for high-throughput whole genome sequencing.
- SOP for testing for the presence of a common Pvmdr1 copy number amplification
- Paper: Genomic Analysis Reveals a Common Breakpoint in Amplifications of the Plasmodium vivax Multidrug Resistance 1 Locus in Thailand.
Genomics resources:
- A new P. vivax reference sequence is available here.
- Paper: A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes.
- Whole genome sequencing data from clinical P. vivax is available here.
- PV4 release coming soon
- Find out more in our genomics publications – see papers below.
Informatics tools:
vivaxGEN-MS
- An online, open-access platform for collating, analyzing and sharing P. vivax microsatellite genotyping data, available at http://vivaxgen.menzies.edu.au/
- Paper: VivaxGEN: An open access platform for comparative analysis of short tandem repeat genotyping data in Plasmodium vivax populations.
vivaxGEN-GEO
- VivaxGen-GEO: A VivaxGEN project: An online, open-access platform with an in-built classifier tool for determining the geographic origin of P. vivax infections using SNP barcodes
- Paper: A molecular barcode and online tool to identify and map imported infection with Plasmodium vivax
vivaxGEN-SNP (in development – available January 2020)
- An online, open-access platform for collating, analysing and sharing P. vivax data generated at Single Nucleotide Polymorphism (SNP) barcodes
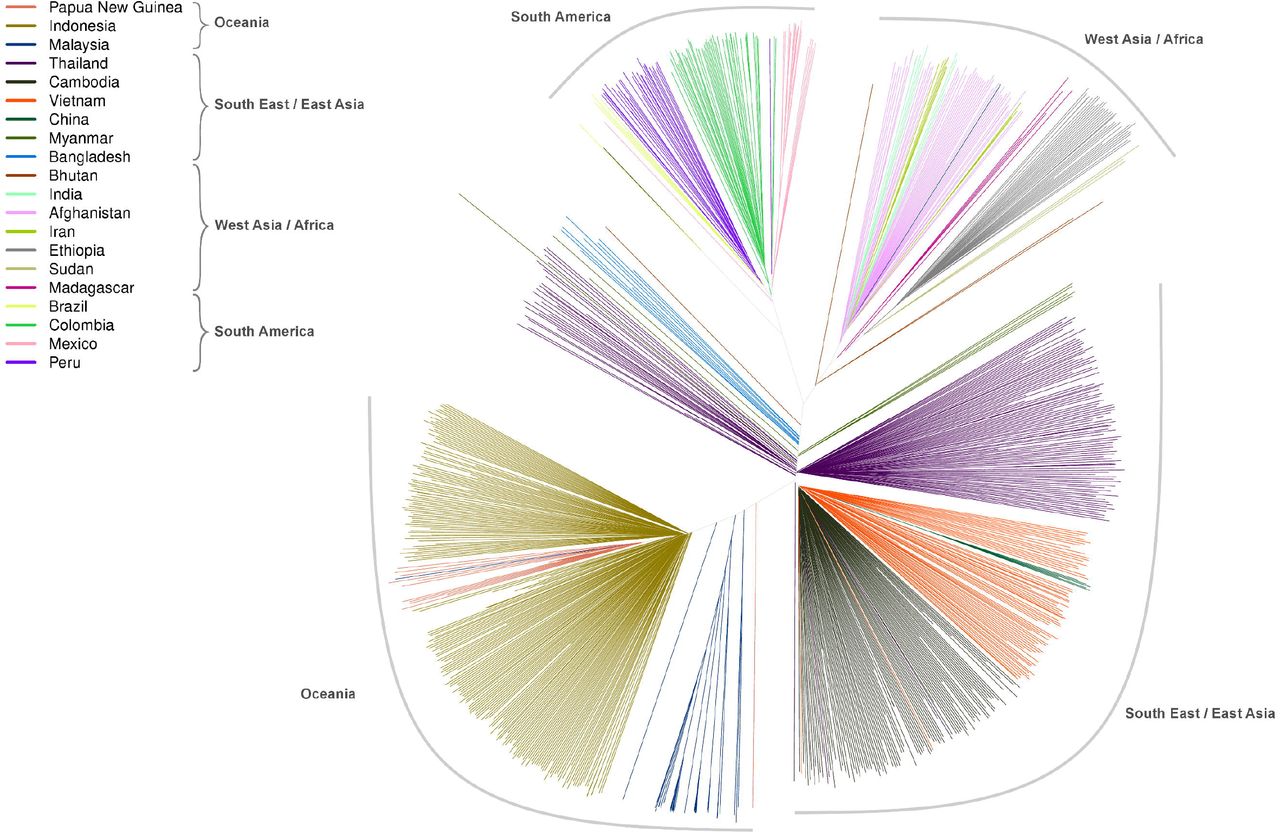
A neighbor-joining tree derived from genomic data on 831 P. vivax infections from 20 countries - Trimarsanto et al., BioRxiv 2019.
Insights on the biology and epidemiology of P. vivax using genomics:
Global
- Trimarsanto et al. (2019). A molecular barcode and online tool to identify and map imported infection with Plasmodium vivax. BioRxiv.
- Pearson et al. (2016). Genomic analysis of local variation and recent evolution in Plasmodium vivax. Nature Genetics, 48(8), 959-964.
- Auburn et al. (2016). A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Research,15,1-4.
Ethiopia
- Auburn et al. (2019). Genomic analysis of Plasmodium vivax in Southern Ethiopia Reveals Selective Pressures in Multiple Parasite Mechanisms. Journal of Infectious Diseases, 220(11), 1738-1749.
Malaysia
- Auburn et al. (2018). Genomic analysis of a pre-elimination Malaysian Plasmodium vivax population reveals selective pressures and changing transmission dynamics. Nature Communications, 9(1), 2585.
Thailand
- Auburn et al. (2016). Genomic analysis reveals a common breakpoint in amplifications of the Plasmodium vivax multidrug resistance 1 locus in Thailand. The Journal of Infectious Diseases, 214(8), 1235-42..
Insights on local parasite transmission dynamics using genotyping:
Bhutan
- Wangchuck et al. (2016). Where chloroquine still works: the genetic make-up and susceptibility of Plasmodium vivax to chloroquine plus primaquine in Bhutan. Malaria Journal, 15(1), 277.
China
- Liu et al. (2014). Genetic diversity and population structure of Plasmodium vivax in Central China. Malaria Journal, 13,262.
Indonesia
- Pava et al. (2017). Passively versus actively detected malaria: similar genetic diversity but different complexity of infection. The American Journal of Tropical Medicine and Hygiene, 97(6), 1788-1796.
- Pava et al. (2017). Genetic micro-epidemiology of malaria in Papua Indonesia: Extensive P. vivax diversity and a subpopulation of asymptomatic P. falciparum infections. PLoS ONE, 12(5), e0177445.
- Noviyanti et al. (2015). Contrasting transmission dynamics of co-endemic Plasmodium vivax and P. falciparum: Implications for malaria control and elimination. PLoS Neglected Tropical Diseases, 9(5), e0003739.
Ethiopia
- Getachew et al. (2015). Variation in complexity of infection and transmission stability between neighboring populations of Plasmodium vivax in southern Ethiopia. PLoS ONE,10(10), e0140780.
Iran
- Hamedi et al. (2016). Molecular epidemiology of P. vivax in Iran: High diversity and complex sub-structure using neutral markers, but no evidence of Y976F mutation at pvmdr1. PLoS ONE, 11(11),e0166124
Republic of Korea
- Kim et al. (2016). Further evidence of increasing diversity of Plasmodium vivax in the Republic of Korea in recent years. PLoS ONE, 11(3), e0151514.
Malaysia
- Abdullah et al. (2013). Plasmodium vivax population structure and transmission dynamics in Sabah Malaysia. PLoS ONE, 8(12):e82553.
Myanmar
- Htun et al. (2017). Chloroquine efficacy for Plasmodium vivax in Myanmar in populations with high genetic diversity and moderate parasite gene flow. Malaria Journal,16(1), 2812017.

In May 2022, A/Prof. Sarah Auburn presented to the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM). This webinar focused on the VivaxGEN framework: Establishing molecular surveillance tools for Plasmodium vivax malaria.
Watch the presentation via Zoom recording. Password required is: #bV1R1wP

At the GenRe-Mekong Scientific Forum in March 2022, A/Prof. Sarah Auburn presented on the use of genetic epidemiology in tackling malaria in the Greater Mekong Sub-regions.
Watch the presentation via YouTube.
Genotyping Workshop
In November 2019, a two-day genotyping workshop co-funded by the Asia Pacific Malaria Elimination Network (APMEN) and Australia’s Department of Foreign Affairs and Trade was conducted at the Eijkman Institute for Molecular Biology in Jakarta, Indonesia. The meeting was attended by 70 participants from nine countries, including 10 National Malaria Control Programs (NMCP), UNICEF and World Health Organisation representatives.
The aim of the meeting was to discuss novel molecular approaches for parasite surveillance and explore ways in which these tools can be integrated into policy and practice.
Find out more about the presentations and discussions on Twitter.
Workshop participants at the Eijkman Institute for Molecular Biology, Jakarta, Indonesia.



